Why DGGS? Motivation#
“A Discrete Global Grid System (DGGS) is a spatial reference system that uses a hierarchical tessellation of cells to partition and address the globe.”
— OGC Abstract Specification, 2017 (OGC 15‑104r5: Topic 21)
Accessible at the Open Geospatial Consortium website oai_citation:0‡docs.ogc.org
Key properties of DGGS:
Equal Area
Seamless Global Coverage
Multi-Scale (hierarchical, supports multiple resolutions)
Traditional map projections introduce distortion:
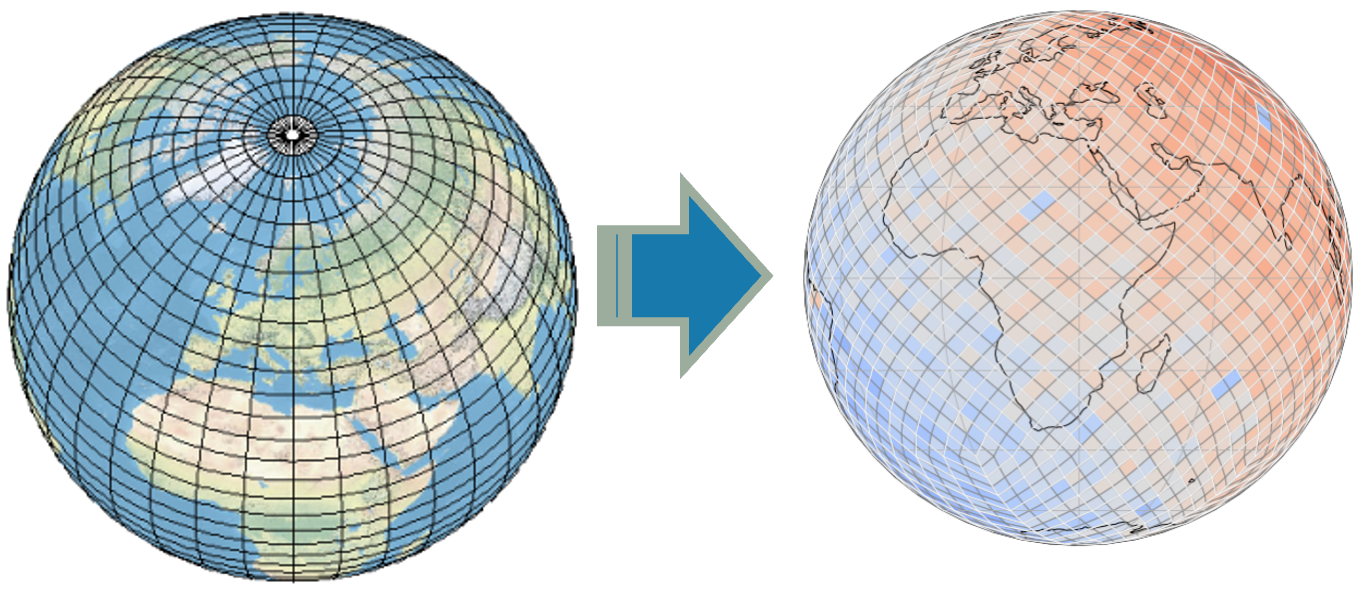
DGGS Families: Examples and Images#
DGGS come in many types. Two of the most widely-used in Earth sciences are H3 and HEALPix:
H3 (Uber)#
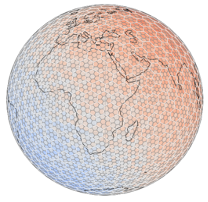
|
HEALPix#
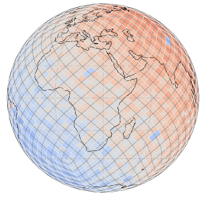
|
|
Both figures are from: | |
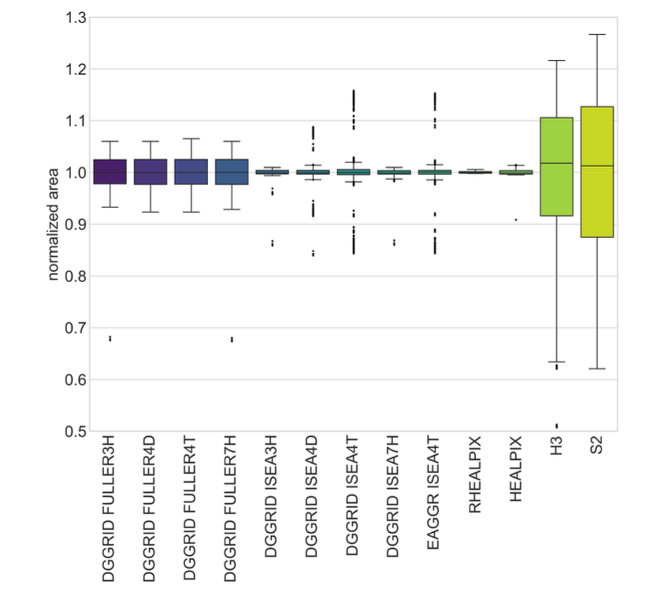
Area Distortion Analysis#
The figure below (also from Kmoch et al., 2024) summarizes normalized area values for various DGGS types, highlighting HEALPix’s strong equal-area properties:
Further resources:
✅ Getting Started: Interactive HEALPix Grid#
Below is an interactive HEALPix grid visualization using Plotly and Healpy.
Each dot represents the center of a HEALPix cell at refinement_level=3 (try changing
refinement_levelfor more or fewer cells).Hover over points to see pixel numbers.
The grid is equal-area and hierarchical, ideal for scalable geospatial analysis.
You can edit and experiment with the code:
Increase
refinement_levelfor higher resolution grids (finer detail).Decrease
refinement_levelfor lower resolution grids (less detail).
For more information, see the HEALPix documentation.
import numpy as np
import healpy as hp
import plotly.graph_objects as go
import plotly.io as pio
import cartopy.io.shapereader as shpreader
from shapely.geometry import LineString, MultiLineString
pio.renderers.default = 'notebook'
def sph2cart(theta, phi):
"""Convert spherical to Cartesian coordinates."""
x = np.sin(theta) * np.cos(phi)
y = np.sin(theta) * np.sin(phi)
z = np.cos(theta)
return x, y, z
def add_coastlines(fig, line_color='gray', line_width=1):
"""Add coastlines to Plotly 3D globe from Cartopy's Natural Earth data."""
reader = shpreader.natural_earth(resolution='110m', category='physical', name='coastline')
geometries = shpreader.Reader(reader).geometries()
for geom in geometries:
lines = [geom] if isinstance(geom, LineString) else list(geom.geoms) if isinstance(geom, MultiLineString) else []
for line in lines:
coords = np.array(line.coords)
if coords.shape[0] < 2:
continue
lons, lats = coords[:, 0], coords[:, 1]
theta, phi = np.radians(90 - lats), np.radians(lons)
x, y, z = sph2cart(theta, phi)
fig.add_trace(go.Scatter3d(
x=x, y=y, z=z,
mode='lines',
line=dict(color=line_color, width=line_width),
showlegend=False
))
def create_healpix_plot(refinement_level=3, show_labels=True, color='red'):
nside = 2**refinement_level
npix = hp.nside2npix(nside)
fig = go.Figure()
# Add globe surface
u, v = np.linspace(0, 2*np.pi, 100), np.linspace(0, np.pi, 100)
x = np.outer(np.cos(u), np.sin(v))
y = np.outer(np.sin(u), np.sin(v))
z = np.outer(np.ones_like(u), np.cos(v))
fig.add_trace(go.Surface(
x=x, y=y, z=z,
colorscale=[[0, 'rgb(80,80,80)'], [1, 'rgb(20,20,20)']],
opacity=1.0,
showscale=False
))
# Plot HEALPix cells
for pix in range(npix):
boundary = hp.boundaries(nside, pix, step=20, nest=True)
xb, yb, zb = boundary
fig.add_trace(go.Scatter3d(
x=np.append(xb, xb[0]),
y=np.append(yb, yb[0]),
z=np.append(zb, zb[0]),
mode='lines',
line=dict(color=color, width=2),
showlegend=False
))
if show_labels:
theta, phi = hp.pix2ang(nside, pix, nest=True)
xc, yc, zc = sph2cart(theta, phi)
fig.add_trace(go.Scatter3d(
x=[xc], y=[yc], z=[zc],
mode='text',
text=[str(pix)],
textfont=dict(size=10, color=color),
showlegend=False
))
# Add coastline outlines
add_coastlines(fig, line_color='#00B5E2', line_width=2.5)
fig.update_layout(
title=f"HEALPix NESTED refinement_level={refinement_level} with Coastlines",
scene=dict(
xaxis=dict(visible=False),
yaxis=dict(visible=False),
zaxis=dict(visible=False),
aspectmode='data'
),
scene_camera=dict(eye=dict(x=1.3, y=1.3, z=1.3)),
margin=dict(l=0, r=0, t=30, b=0)
)
fig.show()
# Example: Plot with refinement_level=3 and labels
create_healpix_plot(refinement_level=1, show_labels=True, color='red')
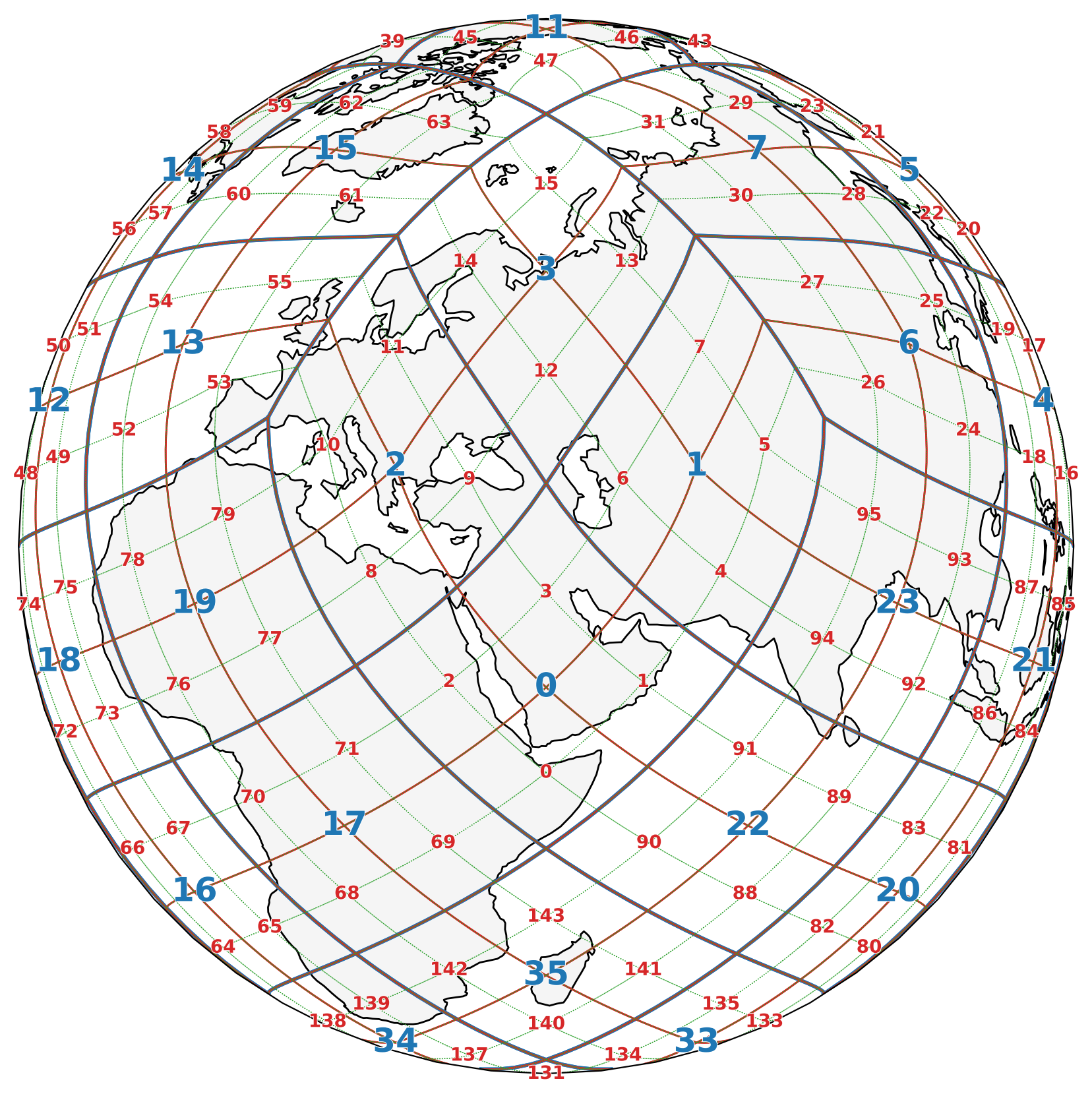
🌐 HEALPix Grid: Parent-Child Connectivity#
The figure below visually demonstrates the hierarchical structure of the HEALPix grid (parent-child connectivity). Different colors represent different refinement levels:
Refinement Level 1 (Parent grid): 🔵 Blue, thicker lines, labeled with pixel IDs.
Refinement Level 2 (Intermediate grid): 🔴 Red, medium lines, labeled with pixel IDs.
Refinement Level 3 (Child grid): 🟢 Green, fine dotted lines, unlabeled for clarity.
This color-coded structure helps highlight how each level nests within the previous, supporting scalable multi-resolution Earth Observation analysis.
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import healpy as hp
import numpy as np
import matplotlib.patheffects as path_effects
def plot_healpix_grid(ax, refinement_level, color='black', linewidth=0.5, linestyle='-',
show_ids=False, text_color='black', fontsize=10):
"""
Plot HEALPix grid lines at a specified refinement level.
"""
nside = 2 ** refinement_level
npix = hp.nside2npix(nside)
for pix in range(npix):
boundary = hp.boundaries(nside, pix, step=10)
lon, lat = hp.vec2ang(boundary.T, lonlat=True)
lon = np.append(lon, lon[0])
lat = np.append(lat, lat[0])
ax.plot(lon, lat, transform=ccrs.Geodetic(),
color=color, linewidth=linewidth, linestyle=linestyle)
if show_ids:
vec = hp.pix2vec(nside, pix,nest=True)
vec = np.array(vec).T
lon_c, lat_c = hp.vec2ang(vec, lonlat=True)
ax.text(lon_c[0], lat_c[0], str(pix),
transform=ccrs.Geodetic(),
fontsize=fontsize,
weight='bold', # Bold font for IDs
ha='center', va='center',
color=text_color,
path_effects=[
path_effects.Stroke(linewidth=0.75, foreground='white'),
path_effects.Normal()
])
# Create figure and globe projection
fig = plt.figure(figsize=(8, 8), dpi=200)
projection = ccrs.Orthographic(central_longitude=45, central_latitude=35)
ax = plt.axes([0, 0, 1, 1], projection=projection)
ax.set_global()
# Earth features
ax.add_feature(cfeature.LAND, facecolor='whitesmoke')
ax.add_feature(cfeature.OCEAN, facecolor='white')
ax.coastlines(resolution='110m')
# Plot: Blue (Level 1), Red (Level 2), Green (Level 3)
plot_healpix_grid(ax, refinement_level=1, color='#1f77b4', linewidth=2, linestyle='-',
show_ids=True, fontsize=18, text_color='#1f77b4') # Blue, bold
plot_healpix_grid(ax, refinement_level=2, color='#d62728', linewidth=1.0, linestyle='-',
show_ids=True, fontsize=10, text_color='#d62728') # Red
plot_healpix_grid(ax, refinement_level=3, color='#2ca02c', linewidth=0.5, linestyle=':',
show_ids=False) # Green
plt.show()
plt.close()
✅ Summary#
Discrete Global Grid Systems (DGGS) provide a powerful, scalable framework for Earth Observation and geospatial data analysis.
You learned:
✅ The limitations of traditional map projections for global-scale, multi-resolution applications (e.g., distortion, seams, lack of hierarchy)
✅ The DGGS and its essential properties:
Equal-area, seamless global coverage, hierarchical/multi-scale access✅ DGGS examples:
H3 (hexagonal, hierarchical, efficient for mobility use cases)
HEALPix (equal-area, iso-latitude, widely used in astronomy and Earth sciences)
Including references and visuals for HEALPix.Understanding refinement levels (multi-resolution)
Exploring parent-child relationships and pixel IDs
🔭 What’s Next?#
🧪 Try applying HEALPix to your own geospatial datasets
🛠️ Explore additional DGGS tools: XDGGS
📚 Dive deeper with Awesome DGGS and Awesome HEALPix
